Protein-Protein Interactions
Building on traditional proximity ligation assay (PLA) technology, Navinci’s Naveni in situ proximity ligation technology (isPLA) uses a fluorescence-based methodology that overcomes common background signal challenges in tissue samples and enables the visualization of low abundance proteins.
Leverage Canopy Multiomic Services and let us handle your next isPLA project, from sample preparation through data acquisition with our CellScape™ Precise Spatial Proteomics platform.

Spatial Expertise
Our scientists have significant expertise in numerous spatial technologies, including IF and IHC, in addition to the spatial platforms in our labs. This breadth of knowledge ensures our dedication to your project success.
Deeper Spatial Data
Combining complementary technologies, like isPLA and CellScape Precise Spatial Proteomics, results in more meaningful and reliable data.
A Partner for your Study
We know how important your time is and how precious your samples are. Our efficient staff will advise on how to get the most out of your samples.
A Comprehensive Portfolio
We offer a specialized mix of spatial and single-cell analysis technologies that complement one another. Ask about how our comprehensive suite of services can provide powerful insights for your study.
NaveniFlex Technology
In situ proximity ligation assays are a well-established tool to detect protein-protein interactions, post-translation modifications, and single protein localization in tissues.
Traditionally, background signal and non-specific fluorphore binding has posed a significant challenge. NaviFlex technology has been optimized to visualize signals that would traditionally be obscured by background, increasing detection sensitivity and specificity for localized proteins and enabling the detection of low abundance proteins. In addition to these innovations, the reagents are designed to be customizable, so researchers can use primary antibodies that bind to their specific targets and are not limited to pre-designed kit availability.
How Our Service Works
Send Us Your Samples
The Navinci kits are suitable for FFPE tissue and fresh frozen tissue. Send us your blocks to section and process. Blocks will be returned upon project completion.
We Do the Work
We use your chosen primary antibodies and use secondary Navenibodies to detect targets near each other, utilizing the CellScape for isPLA image capture. Additional biomarker staining with VistaPlex panels can follow.
We Deliver Your Data
Our scientists provide image data from both isPLA and VistaPlex staining and provide guidance for further in-depth analysis.
How We Approach the Naveni isPLA Workflow
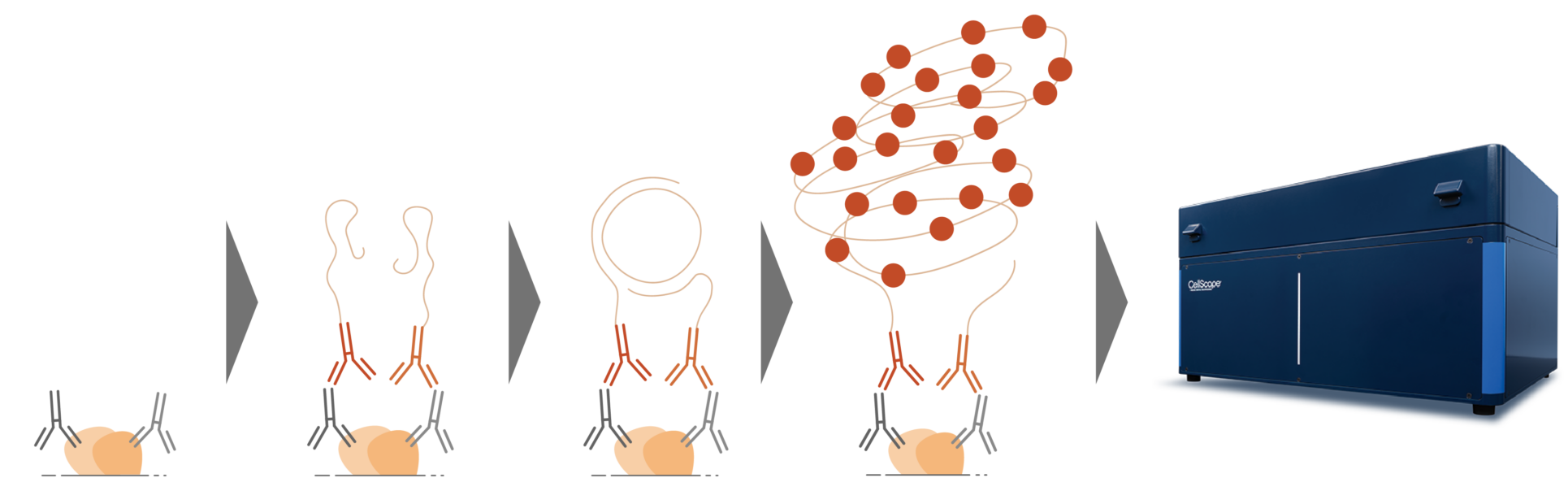
Detection of Protein-Protein Interactions
Primary antibodies bind to their targets and are then recognized by Navenibodies, secondary antibodies conjugated to oligo tags. If the two Navenibodies are in close proximity, their oligo tags serve as templates for rolling circle amplification and subsequent detection by fluorescent probes.
The CellScape Platform
High signal-to-noise ratio with Navinci isPLA enables the detection of individual proximity events. The high-resolution, high dynamic range CellScape platform enables hands-free localization and resolution of individual protein-protein interactions.
Expand Discovery with VistaPlex
Utilizing the Navinci technology on our CellScape™ platform, we can pair protein interaction data with high-plex targeted spatial proteomics. The combination of our VistaPlex™ Multiplex Assay kits following a proximity ligation assay allows for deeper anlysis into the cell types involved in the observed protein-protein interactions. This enables us to analyze the tumor microenvironment in previously unmatched detail.
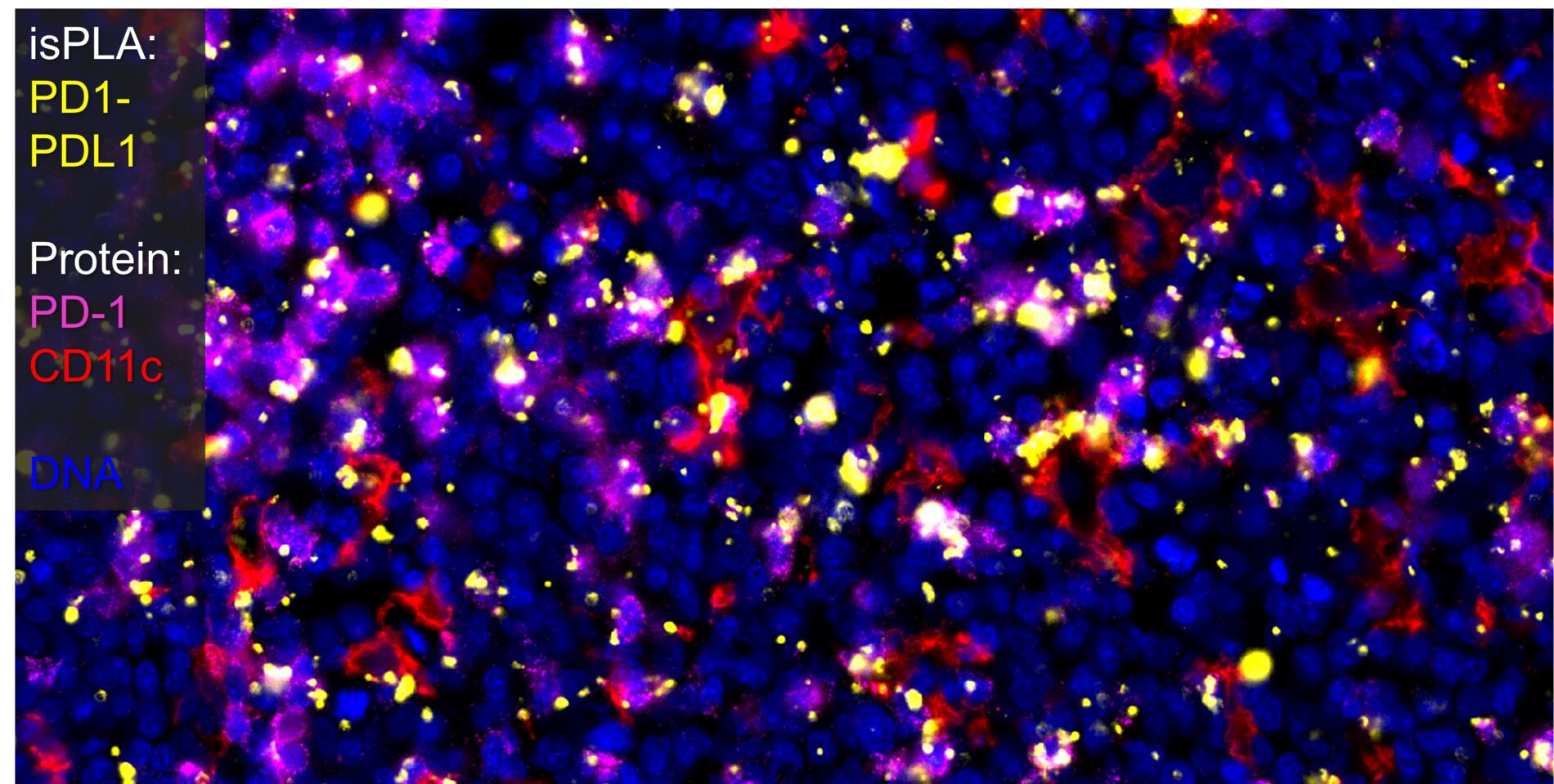
CellScape high resolution imaging allows precise identification of cells involved in PD-1/PD-L1 protein-protein interactions in FFPE tonsil tissue germinal centers.
CellScape Service
We are the exclusive provider of CellScape Precise Spatial Proteomics Services. Leveragd our expertise in biomarker detection, assay development, and quantitative phenotyping for your next spatial biology project.
