Research Spotlight: Mara De Martino Leveraging the CellScape platform and MALDI for Glioblastoma Research
In a recent presentation, Mara De Martino, PhD, a postdoc in Dr. Claire Vanpouille-Box's lab in the Weill Cornell Medicine radiation oncology department, shared her research on radiotherapy-treated glioblastoma, leveraging the CellScape platform and MALDI. Her work, supported in part by Bruker Spatial Biology, delved into the intricate interplay between metabolism and the tumor microenvironment, particularly focused on fatty acid metabolism as a potential immune evasion mechanism in glioblastoma.
Dr. De Martino's study aimed to uncover the impact of radiation on metabolic reprogramming within glioblastoma tumors. By utilizing preclinical models, including injection of tumorigenic cells into mice and use of pharmacological inhibitors, she and her team investigated how radiation-induced metabolic changes influence both tumor progression and immune cell function. Central to her research was the use of advanced omics techniques, notably MALDI-IMS using Bruker’s scimaX® Magnetic Resonance Mass Spectrometry for spatial metabolomics and CellScape™ Precise Spatial Proteomics for targeted spatial proteomics.
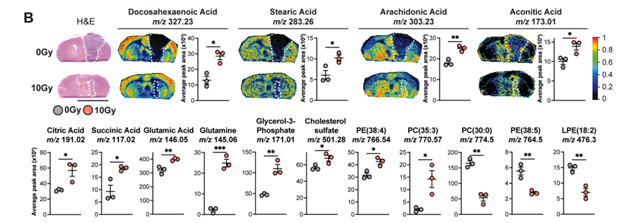
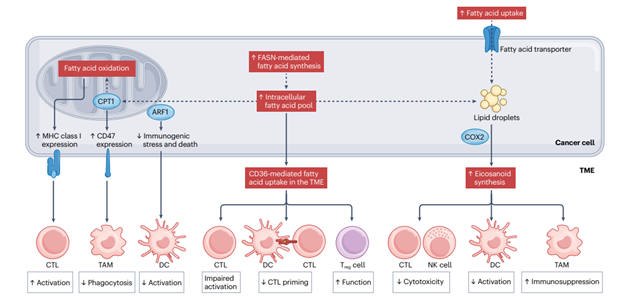
Because radiation-treated tumors are so much smaller than their untreated counterparts, techniques requiring excision of such tiny tumors are prohibitively challenging. Dr. de Martino’s research required a spatial biology approach to better understand gene expression, immune infiltration, and cell-cell interactions in tumors after radiotherapy.
The utilization of CellScape through Canopy Spatial Services allowed Dr. de Martino to visualize and analyze the spatial distribution of various cell populations within a treated glioblastoma tumor. Using multiplex immunofluorescence staining and advanced imaging analysis, she identified immune infiltration and specific immune cell populations within radiation-treated tumors, including dendritic cells, cytotoxic T cells, regulatory T cells, and macrophages.
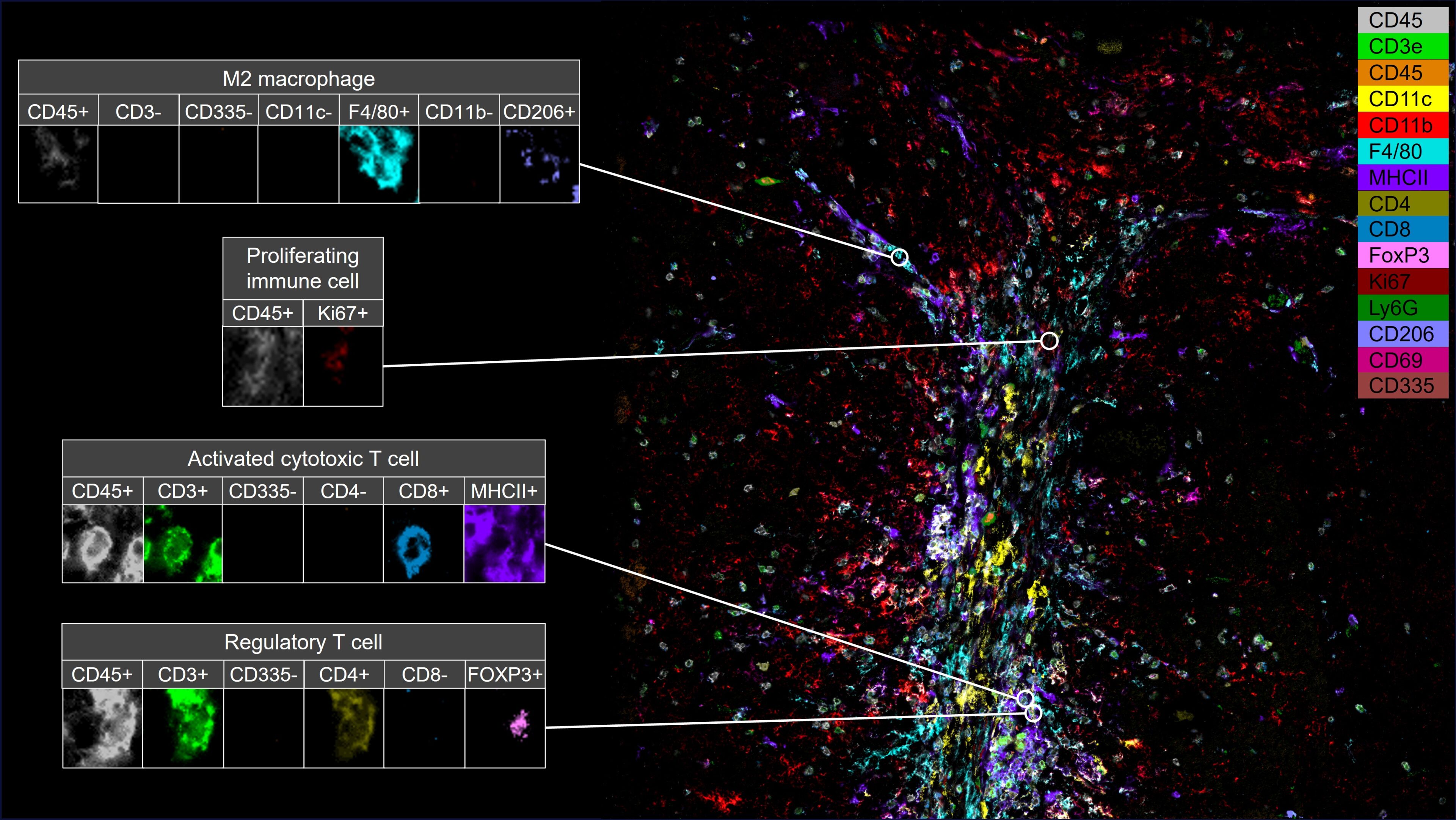
By integrating spatial biology data with metabolic profiling and immune cell analysis, Dr. de Martino uncovered novel insights into the complex mechanisms underlying glioblastoma progression and immune evasion after radiation therapy. Her research sheds light on the potential of targeting fatty acid metabolism as a therapeutic strategy to enhance anti-tumor immune responses and improve treatment outcomes for glioblastoma patients.
Watch the full presentation at IMMUNOLOGY2024 here.
Dr. de Martino's work highlights the invaluable role of spatial biology platforms like CellScape in advancing our understanding of tumor biology and guiding the development of novel therapeutic approaches. Through meticulous spatial analysis, researchers can unravel the intricate interactions between tumor cells, immune cells, and the tumor microenvironment, paving the way for more effective cancer therapies.